What is mRNA?

mRNA is an essential part of you and exists in all of your cells. Your body makes lots of mRNA all the time. mRNA can teach the body how to make its own medicines in the form of proteins.

Each mRNA carries instructions like a "blueprint" to make a specific protein. Cells interpret this blueprint and put the protein together. Antibodies are one type of protein that help your body fight diseases.

mRNA is powerful but fragile. mRNA stability needs to be carefully optimized to maximize in-cell protein production and the shelf life of the drug product. Once mRNA does its job, it doesn't stay around for very long and is broken down by the body.
visit aboutmrna.com
Computational Challenges of mRNA Design
Secondary Structure Prediction
Accurate prediction of mRNA's secondary structure is vital for its functionality in protein synthesis. Advanced computational models are essential for designing mRNA sequences that not only fold as desired but also maintain high stability under physiological conditions.
Importance of Optimization
Optimizing mRNA for therapeutic use involves a delicate balance of stability, efficient protein translation, and minimal immune response. Advanced computational tools and bioinformatics are utilized to enhance these properties, aiding in the development of effective and safe mRNA-based therapies.
How can Quantum Computing accelerate mRNA therapeutic design?
Structure Defines Success
Explore More,
Discover Better
More Diversity,
Better Outcomes
- Accurate mRNA structure prediction is crucial for optimal folding, ensuring stability and function, but is computationally intensive for classical systems which often rely on approximations to limit possible secondary structures.
- Quantum computing promises to lift some of these limitations by more effectively exploring the solution space, potentially revolutionizing our approach to mRNA design.
- This task is an NP-complete problem, challenging traditional computational methods with its complexity.
- Efficient exploration of vast mRNA sequence spaces unveils novel, more diverse, and stable therapeutic mRNA designs.
- Quantum algorithms evaluate multiple design candidates simultaneously, accelerating search and improving the selection of effective sequences.
- Quantum computing has the potential to accelerate mRNA drug discovery by enhancing the efficiency of mRNA optimization.
- Broader diversity of mRNA sequences enables discovery of higher-quality mRNA designs with optimal therapeutic properties.
- Quantum computing could enable rapid evaluation of diverse molecular configurations, enhancing the efficiency of design processes.
- Enhanced sampling methods made possible by quantum computing may improve the quality of the final therapeutic candidates.
Building blocks of mRNA Secondary Structure
mRNA structure prediction involves calculating the most stable configuration of nucleotide sequences. The lowest energy folding is indicative of the molecule's equilibrium state, crucial for its biological function.
To determine the lowest energy conformation, nearest-neighbor rules are utilized. These rules evaluate the individual energy contributions from secondary structure motifs like loops, stems, and bulges, which collectively determine the total energy function and predict the most probable folding of the mRNA.
The number of possible secondary structures an mRNA can adopt is estimated as 2.3n, where n is the length of the sequence. A typical 2,000-nucleotide therapeutic mRNA can theoretically assume an astronomical number of structures.
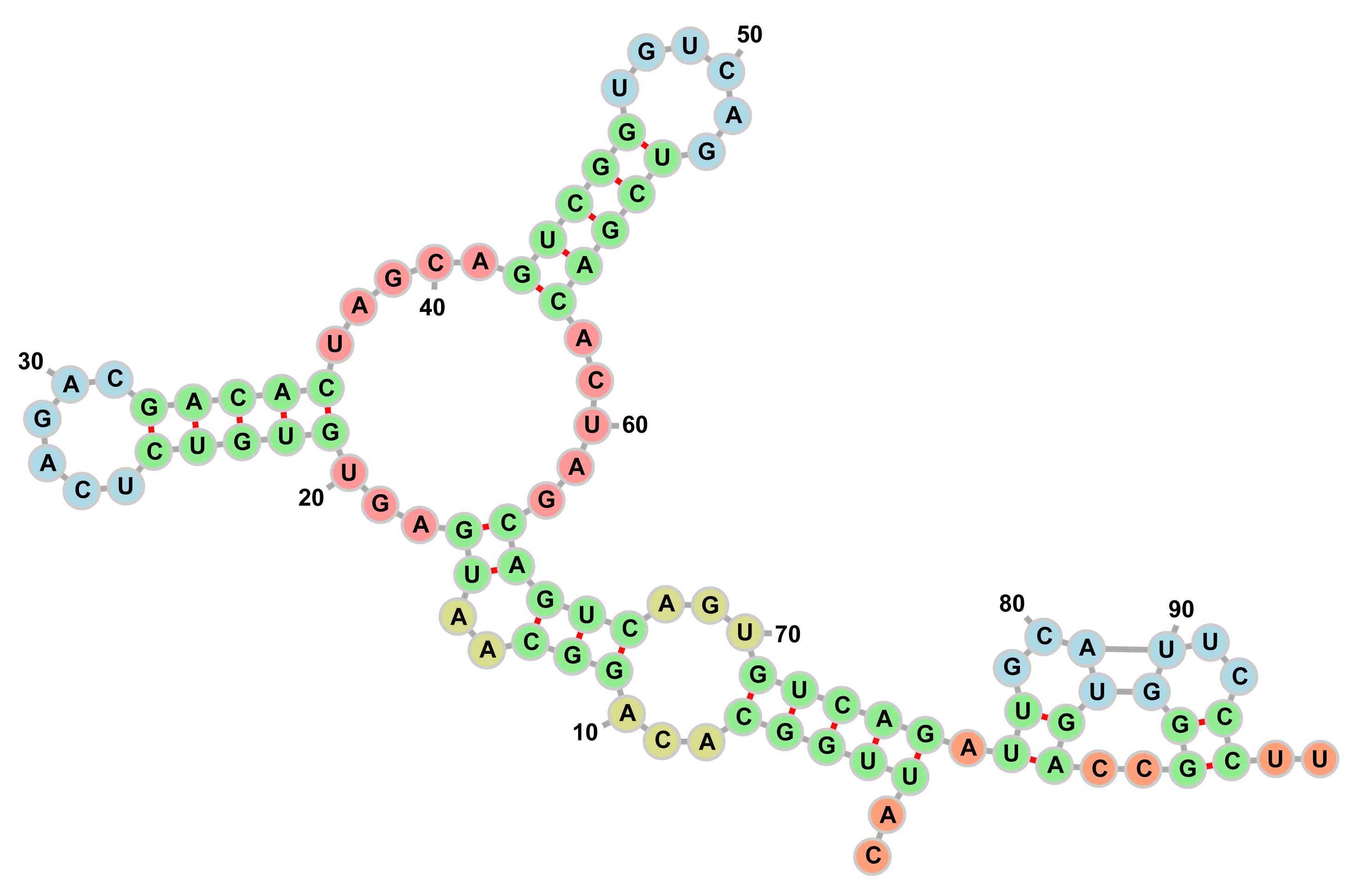
Demo
In this demo, we will demonstrate the results of our quantum algorithm for secondary structure prediction for a range of mRNA sequences solved on state-of-the-art utility-scale quantum computers.
Select the mRNA sequence from the drop-down list and review the algorithm parameters and the problem instance details. The image on the right will show the mRNA sequence before and after optimization.
| Sequence | |
|---|---|
| Length | |
| Number of Constraints | |
| QPU Backend | |
| Number of Qubits | |
| Normalized Energy |
Conclusions and Outlook
- Accurately predicted mRNA secondary structures with up to 45 nucleotides on utility-scale quantum processors with over 100 qubits.
- Laid the groundwork for future research in quantum-assisted bioinformatics, highlighting the importance of developing scalable quantum algorithms for real-world biological problems.
- In the future, it is essential to address technical and algorithmic challenges to scale up the method and approach the problem sizes required for commercial applications.
